How to Build Medical Imaging Workflows Using Cloud and AI
Introduction: How to Build Medical Imaging Workflows Using Cloud and AI
How to Build Medical Imaging Workflows Using Cloud and AI. This guide explains why cloud and AI now power modern imaging pipelines. Cloud services provide scalable storage and compute. AI models speed up image triage, segmentation, and quantification. For developers and designers, this combination reduces time to market. For business owners, it improves ROI through automation and integration. In New Zealand, teams must consider the Privacy Act 2020 and local data residency. Moreover, clinicians expect responsive web interfaces and reliable image access. Therefore, you should design for performance, security and UX from the start. This article suits freelancers, web developers, designers and small teams. It covers core concepts, tooling, code examples and real-world patterns. Finally, you will see checklists and next steps to prototype a production-ready MedTech workflow.
The Foundation of Medical Imaging Workflows Using Cloud and AI
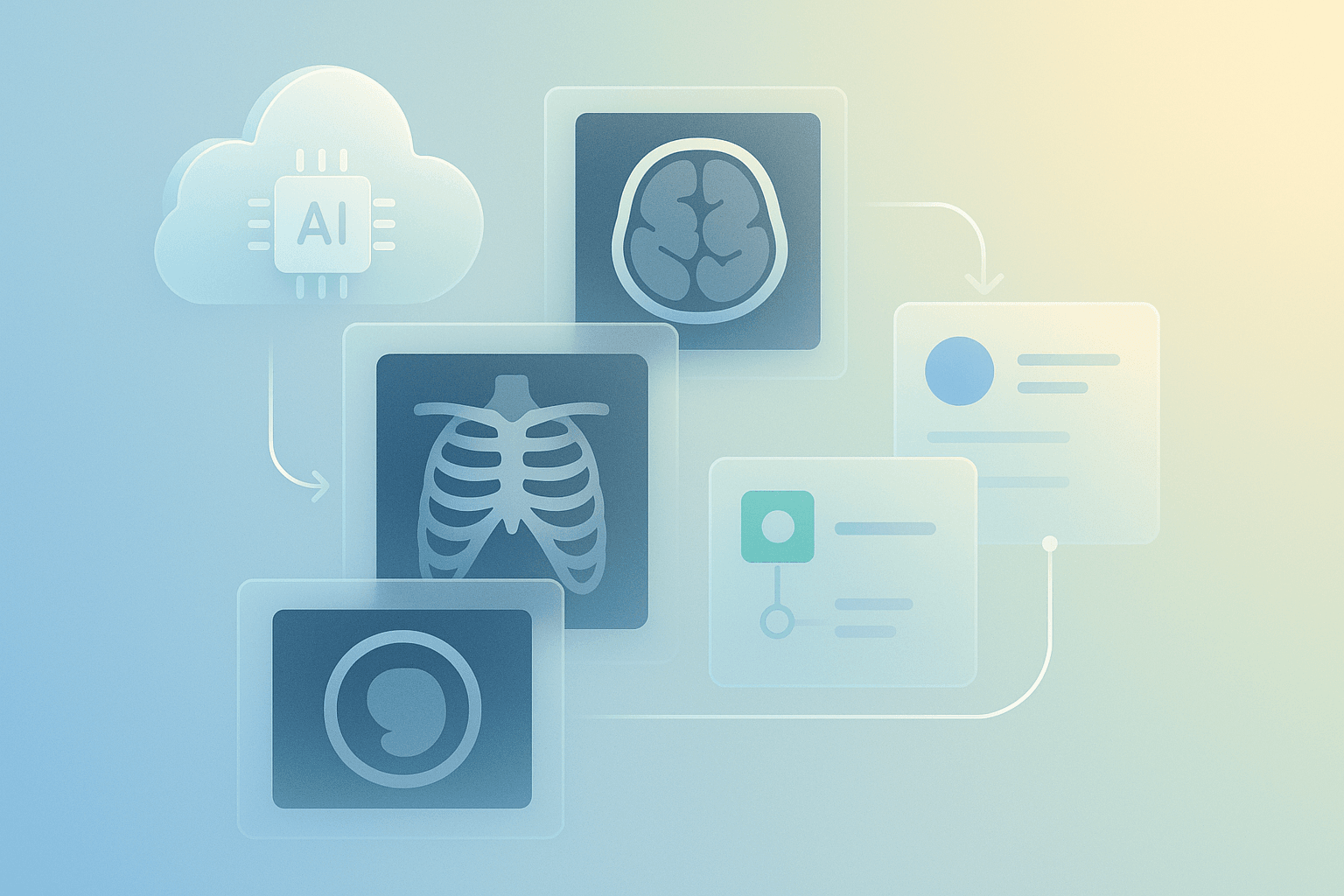
Start with the basics. A robust workflow has four layers: ingestion, storage, processing and presentation. Ingestion handles modalities and formats, typically DICOM or converted NIfTI. Storage covers object stores and specialised PACS. Processing runs AI inference and transforms images. Presentation serves DICOMweb, FHIR metadata and web viewers. In addition, logging, auditing and access control underpin trust. For interoperability, choose DICOMweb and HL7 FHIR wherever possible. Use containerised services and orchestration for reliability. Also, select encryption at rest and in transit, both required for clinical use. From an architecture view, prefer microservices for inference and serverless for bursts. Moreover, design APIs to be stateless and idempotent. This foundation reduces vendor lock-in and makes future integrations simpler and cheaper.
Configuration and Tooling
Choose proven tools first. First of all, for PACS and DICOM services, use Orthanc or dcm4che. Secondly, for AI frameworks, pick MONAI, PyTorch or TensorFlow. Third, for model serving, use NVIDIA Triton or TF Serving. Fourth, for cloud providers, consider AWS, Google Cloud and Azure. In NZ, you may prefer nearby regions like Sydney for lower latency, or a provider with NZ data residency. Fifth, for CI/CD and infra use Terraform, GitHub Actions and Argo CD. Sixth, for observability, pick Prometheus, Grafana and Elastic. Seventh, for prototyping UI and UX, use Figma or Adobe XD. Finally, integrate DICOMweb viewers such as OHIF to reduce front-end effort and speed up demos.
Development and Customisation for Medical Imaging Workflows Using Cloud and AI
Begin with a minimum viable pipeline. First, ingest DICOM files into object storage. Next, run a lightweight container to extract metadata. Then trigger AI inference on a GPU node or serverless endpoint. Finally, return results to the viewer with annotations and structured reports. Below is a minimal Python example using pydicom and AWS S3 as storage. Replace keys with your secrets and use IAM roles for production.
import boto3
from pydicom import dcmread
s3 = boto3.client('s3')
obj = s3.get_object(Bucket='my-bucket', Key='studies/1/image1.dcm')
with open('/tmp/image.dcm', 'wb') as f:
f.write(obj['Body'].read())
ds = dcmread('/tmp/image.dcm')
print(ds.PatientName, ds.Modality)Next, expose an inference endpoint. Use Docker and Triton for GPU throughput. For front-ends, implement secure upload and progressive loading. For example, use this JavaScript pattern to POST DICOM in the browser:
async function uploadDICOM(file){
const fd = new FormData();
fd.append('file', file);
const res = await fetch('/api/upload', { method:'POST', body:fd });
return res.json();
}Also, add caching headers and range requests to improve viewer performance. Finally, instrument latency metrics and autoscaling to control costs while maintaining responsiveness.
Development and Customisation: Design, Performance and Integration
Design principles matter. Prioritise clarity, minimal clicks and contextual information. Use layered UI panels and soft contrast for radiologists. Also, allow toggling of AI overlays and confidence scores. For performance, prefer streaming tiles and progressive rendering. Use CDNs for static assets and edge caching for patient images where policy allows. In addition, profile model latency and use batching in Triton to boost throughput. Use GPU instances sparingly to manage costs and scale with Kubernetes autoscalers. For integration, map model outputs to FHIR DiagnosticReport resources. Moreover, implement role-based access control and audit trails. For ROI, highlight time savings, reduced read times and improved throughput in demos. Finally, produce a prototype that is portfolio-ready and includes clear documentation for clinicians and engineers.
Real-World Examples / Case Studies
Here are three compact use cases that show practical outcomes. First, a regional imaging centre in Australia moved archive storage to object storage. They used a serverless pipeline for anonymisation and saved 40% on storage costs. Second, a NZ tele-radiology start-up used Triton with MONAI models hosted in a managed Kubernetes cluster. They reduced triage time by 60% and improved referral turnaround. Third, a hospital integrated an AI nodule detection model into their PACS through DICOMweb and FHIR. Radiologists accepted the workflow because overlays were non-intrusive and confidence indicators were clear. For prototypes, present screenshots and interactive viewers. Use anonymised datasets and demonstrate performance under simulated load. Also measure engagement metrics and quantify reduced time per case for ROI conversations.
Checklist
Use this QA list before shipping.
- Confirm legal compliance with the NZ Privacy Act and, if applicable, Ministry of Health guidelines.
- Verify data residency and encryption for stored images.
- Validate DICOM conformance with Orthanc or dcm4che.
- Test AI models for bias, accuracy and edge cases.
- Load-test viewers with synthetic traffic and measure latency.
- Implement CI/CD for model and infra changes using Terraform and GitHub Actions.
- Add logging, metrics and alerting with Prometheus and Grafana.
- Document APIs and provide a clinician-facing guide.
- Ensure disaster recovery with cross-region backups and versioned storage.
- Plan cost controls, autoscaling and rightsizing for GPUs.
Follow these steps to reduce risk and speed time to market.
Key takeaways
Key takeaways: first, combine cloud and AI to scale imaging workflows; second, choose open standards like DICOMweb and FHIR; third, use Orthanc, MONAI and Triton for faster delivery; fourth, design for UX, performance and NZ compliance; finally, measure ROI with throughput and time-saved metrics.
Conclusion
Now you have a practical roadmap. Start small with a prototype that ingests images, runs inference and displays results. Next, iterate with clinicians and collect feedback. For production, hardened security, audits and CI/CD pipelines. If you need hosting in New Zealand, check regional availability and privacy requirements. Spiral Compute Limited can help with architecture review, managed deployment and NZ compliance advice. Finally, build a measurable pilot, quantify benefits and scale confidently. Reach out to partners, use prototyping tools like Figma, and show clinicians a responsive, clear demo. Good luck building your medical imaging workflow.